GEOS-CHEM5¶
The GEOS-CHEM package provides a sasktran.Climatology and a ISKClimatology interface to the GEOS-CHEM 0.625 degree model output stored on the NSCC data portal .
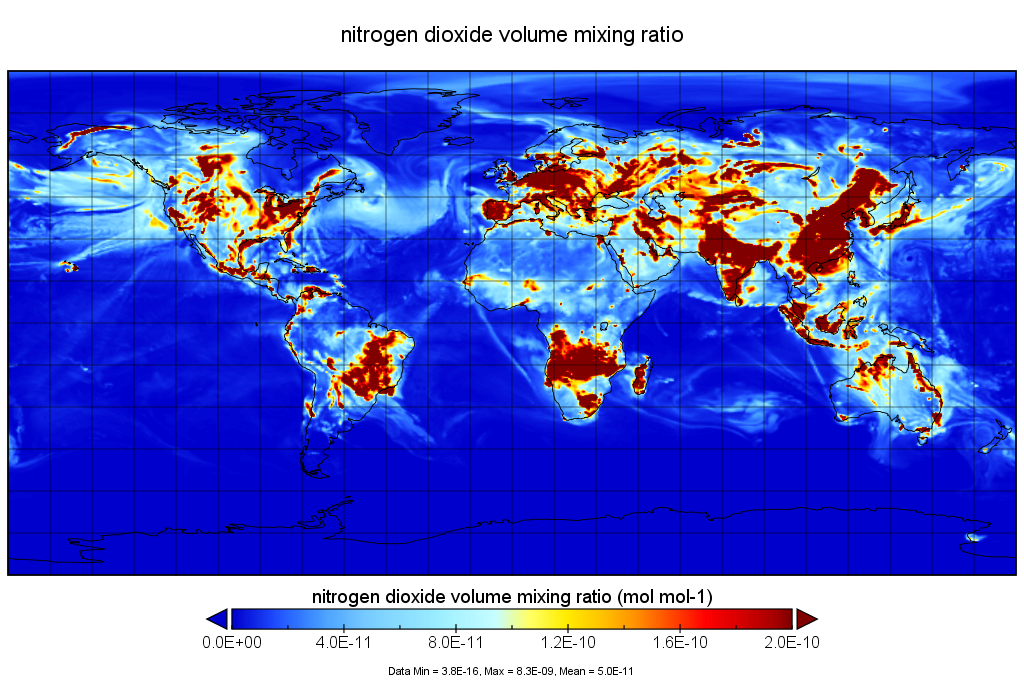
Quick Start¶
Here is an example that shows you how to use GEOS-CHEM. With a bit of luck this will be all you really need. If you need more info then you will have to read the rest of the documentation:
from datetime import datetime
import numpy as np
import matplotlib.pyplot as plt
from sasktran_gcm import GEOS_CHEM
climate = GEOS_CHEM() # Create the GEOS-CHEM object
climate.add_species ( 'SKCLIMATOLOGY_NO2_CM3') # Tell GEOS-CHEM which variables you want to load. You must do this before calling UpdateCache.
climate.add_species ( 'SKCLIMATOLOGY_NO2_VMR') # Add all the variables you want. Each variable added will require several seconds of processing in UpdateCache
latitude=50.3456289 # Set up a reference location
longitude=247.982739087 # The latitude, longitude and utc
utc=datetime.fromisoformat('2013-09-21T12:00:00.000000') # should be "inside" the GEOS-CHEM regional output ranges.
climate.update_cache(latitude,longitude, utc) # Load up the cache. The time variable is used to load in two snapshots that straddle the location mjd
# All variables are now fixed. Only variables specified in `addvariable` are available
h = np.arange( 0, 80000.0, 100.0) # Prepare to extract profiles. Create a height resolution set of heights, 0 to 60 km
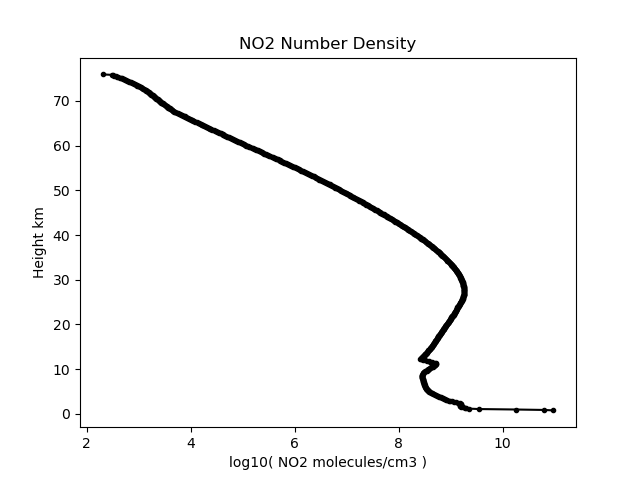
no2cm3 = climate.get_parameter( 'SKCLIMATOLOGY_NO2_CM3', latitude, longitude, h, utc) # Get a height profile of NO2 number density at the given location
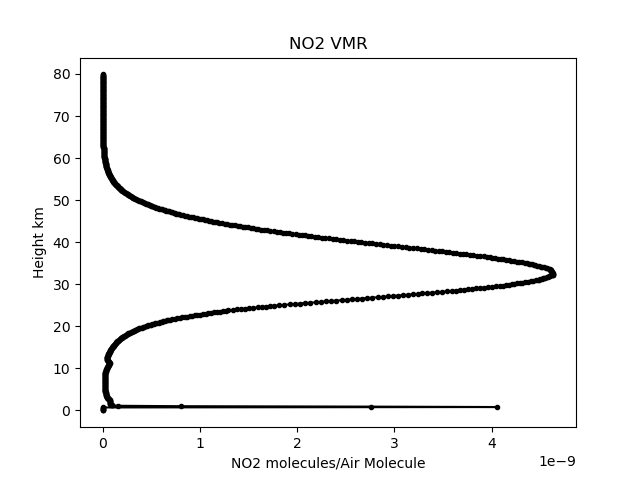
no2vmr = climate.get_parameter( 'SKCLIMATOLOGY_NO2_VMR', latitude, longitude, h, utc) # Get a height profile of NO2 volume mixing ratio at the given location
plt.figure(1) # and plot the data plot the data.
plt.plot( no2vmr,h/1000.0, 'k.-')
plt.title('NO2 VMR')
plt.ylabel('Height km')
plt.xlabel('NO2 molecules/Air Molecule')
plt.figure(2)
plt.plot( np.log10(no2cm3),h/1000.0, 'k.-')
plt.title('NO2 Number Density')
plt.ylabel('Height km')
plt.xlabel('log10( NO2 molecules/cm3 )')
plt.show()
print('Done')
Sasktran Usage¶
The Sasktran GEOS-CHEM climatology is developed in C++ and is compliant with the ISKClimatology interface within the SasktranIF framework. A wrapper class has been developed for use with the Sasktran API:
>>> from sasktran_gcm import GEOS_CHEM
>>> gcm = GEOS_CHEM()
>>> print(gcm)
SasktranIF Climatology: GEOS-CHEM
Supported Species: ['SKCLIMATOLOGY_AIRNUMBERDENSITY_CM3', 'SKCLIMATOLOGY_PRESSURE_PA', 'SKCLIMATOLOGY_TEMPERATURE_K', 'SKCLIMATOLOGY_EPV',
'SKCLIMATOLOGY_GEOMETRIC_HEIGHT', 'SKCLIMATOLOGY_SURFACE_GEOPOTENTIAL_HEIGHT', 'SKCLIMATOLOGY_SURFACE_GEOMETRIC_HEIGHT',
'SKCLIMATOLOGY_SURFACE_PRESSURE_PA', 'SKCLIMATOLOGY_CLOUD_FRACTION', 'SKCLIMATOLOGY_QI_MMR', 'SKCLIMATOLOGY_QL_MMR',
'SKCLIMATOLOGY_QV', 'SKCLIMATOLOGY_RH', 'SKCLIMATOLOGY_AOA_DAYS', 'SKCLIMATOLOGY_DUST_0p7mu', 'SKCLIMATOLOGY_DUST_1p4mu',
'SKCLIMATOLOGY_DUST_2p4mu', 'SKCLIMATOLOGY_DUST_4p5mu', 'SKCLIMATOLOGY_BCPI', 'SKCLIMATOLOGY_BCPO', 'SKCLIMATOLOGY_SALA',
'SKCLIMATOLOGY_SALC', 'SKCLIMATOLOGY_OCPI', 'SKCLIMATOLOGY_OCPO', 'SKCLIMATOLOGY_BRO_CM3', 'SKCLIMATOLOGY_BRO_VMR',
'SKCLIMATOLOGY_C3H6O_CM3', 'SKCLIMATOLOGY_C3H6O_VMR', 'SKCLIMATOLOGY_CH3I_CM3', 'SKCLIMATOLOGY_CH3I_VMR',
'SKCLIMATOLOGY_C5H8_CM3', 'SKCLIMATOLOGY_C5H8_VMR', 'SKCLIMATOLOGY_CH2O_CM3', 'SKCLIMATOLOGY_CH2O_VMR', 'SKCLIMATOLOGY_CH4_CM3',
'SKCLIMATOLOGY_CH4_VMR', 'SKCLIMATOLOGY_CO_CM3', 'SKCLIMATOLOGY_CO_VMR', 'SKCLIMATOLOGY_HNO2_CM3', 'SKCLIMATOLOGY_HNO2_VMR',
'SKCLIMATOLOGY_HNO3_CM3', 'SKCLIMATOLOGY_HNO3_VMR', 'SKCLIMATOLOGY_NH3_CM3', 'SKCLIMATOLOGY_NH3_VMR', 'SKCLIMATOLOGY_NITS',
'SKCLIMATOLOGY_NO2_CM3', 'SKCLIMATOLOGY_NO2_VMR', 'SKCLIMATOLOGY_NO_CM3', 'SKCLIMATOLOGY_NO_VMR', 'SKCLIMATOLOGY_O3_CM3',
'SKCLIMATOLOGY_O3_VMR', 'SKCLIMATOLOGY_PAN_CM3', 'SKCLIMATOLOGY_PAN_VMR', 'SKCLIMATOLOGY_SO2_CM3', 'SKCLIMATOLOGY_SO2_VMR',
'SKCLIMATOLOGY_SO4_CM3']
>>>
GEOS_CHEM Class¶
-
class
sasktran_gcm.geoschem_climatology.GEOS_CHEM¶ A climatology for the GEOS-CHEM model. The code works with the netcdf4 files archived at the NSCC data portal.
Examples
>>> from sasktran_gcm import GEOS_CHEM >>> geos = GEOS_CHEM() >>> print(geos) SasktranIF Climatology: GEOS-CHEM Supported Species: ['SKCLIMATOLOGY_AIRNUMBERDENSITY_CM3', 'SKCLIMATOLOGY_PRESSURE_PA', 'SKCLIMATOLOGY_TEMPERATURE_K', 'SKCLIMATOLOGY_EPV', 'SKCLIMATOLOGY_GEOMETRIC_HEIGHT', 'SKCLIMATOLOGY_SURFACE_GEOPOTENTIAL_HEIGHT', 'SKCLIMATOLOGY_SURFACE_PRESSURE_PA', 'SKCLIMATOLOGY_CLOUD_FRACTION', 'SKCLIMATOLOGY_QI_MMR', 'SKCLIMATOLOGY_QL_MMR', 'SKCLIMATOLOGY_QV', 'SKCLIMATOLOGY_RH', 'SKCLIMATOLOGY_AOA_DAYS', 'SKCLIMATOLOGY_DUST_0p7mu', 'SKCLIMATOLOGY_DUST_1p4mu', 'SKCLIMATOLOGY_DUST_2p4mu', 'SKCLIMATOLOGY_DUST_4p5mu', 'SKCLIMATOLOGY_BCPI', 'SKCLIMATOLOGY_BCPO', 'SKCLIMATOLOGY_SALA', 'SKCLIMATOLOGY_SALC', 'SKCLIMATOLOGY_OCPI', 'SKCLIMATOLOGY_OCPO', 'SKCLIMATOLOGY_BRO_CM3', 'SKCLIMATOLOGY_BRO_VMR', 'SKCLIMATOLOGY_C3H6O_CM3', 'SKCLIMATOLOGY_C3H6O_VMR', 'SKCLIMATOLOGY_CH3I_CM3', 'SKCLIMATOLOGY_CH3I_VMR', 'SKCLIMATOLOGY_C5H8_CM3', 'SKCLIMATOLOGY_C5H8_VMR', 'SKCLIMATOLOGY_CH2O_CM3', 'SKCLIMATOLOGY_CH2O_VMR', 'SKCLIMATOLOGY_CH4_CM3', 'SKCLIMATOLOGY_CH4_VMR', 'SKCLIMATOLOGY_CO_CM3', 'SKCLIMATOLOGY_CO_VMR', 'SKCLIMATOLOGY_HNO2_CM3', 'SKCLIMATOLOGY_HNO2_VMR', 'SKCLIMATOLOGY_HNO3_CM3', 'SKCLIMATOLOGY_HNO3_VMR', 'SKCLIMATOLOGY_NH3_CM3', 'SKCLIMATOLOGY_NH3_VMR', 'SKCLIMATOLOGY_NITS', 'SKCLIMATOLOGY_NO2_CM3', 'SKCLIMATOLOGY_NO2_VMR', 'SKCLIMATOLOGY_NO_CM3', 'SKCLIMATOLOGY_NO_VMR', 'SKCLIMATOLOGY_O3_CM3', 'SKCLIMATOLOGY_O3_VMR', 'SKCLIMATOLOGY_PAN_CM3', 'SKCLIMATOLOGY_PAN_VMR', 'SKCLIMATOLOGY_SO2_CM3', 'SKCLIMATOLOGY_SO2_VMR', 'SKCLIMATOLOGY_SO4_CM3']
-
add_species(speciesname)¶ Instructs the GEOS-CHEM climatology to load the requested variable when it next loads the internal cache. The function may be called multiple times to request multiple variables be loaded as long as all the calls are made before the first call to update_cache or get_parameter.
- Parameters
speciesname (str) – The name of any species supported by GEOS-CHEM. Complete list of species can be found in method supportedspecies of in the GEOS-CHEM documentation.
- Returns
Returns True if successful.
- Return type
bool
-
force_year_to_use(year)¶ Forces GEOS-CHEM to load data files from the specified year regardless of the actual year requested by the user calculations. This is useful to allow the code to use GEOS CHEM data from one year even when performing calculations for another year. The year parameter actually runs from July of the given year to June of the next year, so specifying 2013 will force GEOS CHEM to load files from July of 2013 to June of 2014 regardless of the year entered by the user. Only the year field is modified and it is only applied to file I/O operations.
- Parameters
year (int) – The year of the files which will be loaded from file. The year runs from July of the specified year to June of the next year
- Returns
Returns True if successful.
- Return type
bool
-
get_parameter(species: str, latitude: float, longitude: float, altitudes: numpy.ndarray, utc: float) → numpy.ndarray¶ Get a profile from the climatology. The code will update the internal cache if necessary. You must make all the necessary calls to add_species before the first call to get_parameter. Requesting to add species after the first call to update_cache or get_parameter is not supported.
- Parameters
species (str) – Species identifier of the species to query. Corresponds to
sasktran.Species.species. The species must have been requested by a previous call to load_specieslatitude (float) – Latitude in degrees.
longitude (float) – Longitude in degrees.
altitudes (np.ndarray) – Profile altitude grid in meters.
mjd (float) – A scalar UTC that can be converted to Modified julian date. It can be a datetime, datetime64 or (mjd) number
- Returns
Climatology values at the specified altitudes.
- Return type
np.ndarray
-
supported_species()¶ Get a list of species identifiers that are supported by this climatology.
- Returns
Species identifiers that are supported by this climatology. Identifiers correspond to
sasktran.Species.species- Return type
list
-
update_cache(latitude: float, longitude: float, utc: float) → bool¶ Updates the GEOS-CHEM internal cache. Once this function is successfully called the cache will stay frozen in memory until the next call to update_cache. This can be useful to avoid constant swapping of the cache. The GEOS-CHEM cache is currently only updated if the UTC goes out of range of the current cache.
The user must make all the necessary calls to add_species before calling update_cache as only the species tagged for loading will be loaded into the cache.
- Parameters
latitude (float) – Latitude in degrees.
longitude (float) – Longitude in degrees.
mjd (float) – A scalar UTC that can be converted to Modified julian date. It can be a datetime, datetime64 or (mjd) number
- Returns
True if cache is successfully loaded.
- Return type
bool
-
SasktranIF Property List¶
Users may wish to program the geos-chem climatology directly through the SasktranIF interface:
ok = gcm.skif_object.SetProperty('PropertyName', value)
The property name is not case sensitive, and the value may be a scalar, list, or array depending upon context
-
add_species(stringvariablename)¶ Specifies the name of a species to load at the next call to UpdateCache. Multiple variables can be requested using multiple calls to add_species but all calls must be made before the first call to update_cache or get_parameter.
-
force_year_to_use(double year_to_load)¶ Forces the climatology to load data files from the specified year regardless of the actual year requested by the user calculations. This is useful to allow the code to use GEOS CHEM data from one year even when performing calculations for another year. The year_to_load parameter actually runs from July of the given year to June of the next year , so specifying 2013 will force GEOS CHEM to load files from July of 2013 to June of 2014 regardless of the year entered by the user. Only the year field is modified and it is only applied to file I/O operations.
Supported Species¶
The sasktran GEOS-CHEM package supports all the species stored as Geos-Chem output variables in the NCCS online data portal. Each variable is given at a specific UTC and specifies a 3-D grid spanning the entire globe consisting of 576 longitude points spanning -180 to +180 degrees longitude, 361 points spanning -90 to +90 degrees latitude and 72 sigma pressure levels. Each snapshot of each Geos-Chem variable occupies around 120 MB of memory and the climatology always loads in the two snapshots that straddle the time of interest, requiring a total of 240 MB of memory for each variable.
The table below identifies the geophysical parameters supported by the GEOS-CHEM package as well as the source files that must be loaded on to users system. Note that the Sasktran climatology values may be derived from several GEOS-CHEM variables. For example, all number density parameters must use the corresponding GEOS-CHEM VMR variable and well as pressure and temperature.
Sasktran Handle
Description
Geos-Chem File/Variable
SKCLIMATOLOGY_AIRNUMBERDENSITY_CM3
Air number density
ASM PL and T
SKCLIMATOLOGY_PRESSURE_PA
Pressure
ASM PL
SKCLIMATOLOGY_TEMPERATURE_K
Temperature
ASM T
SKCLIMATOLOGY_EPV
Ertels Potential Vorticity
ASM EPV
SKCLIMATOLOGY_GEOMETRIC_HEIGHT
Geometric height.
ASM H
SKCLIMATOLOGY_SURFACE_GEOPOTENTIAL_HEIGHT
Surface Geopotential Height (meters)
ASM PHIS
SKCLIMATOLOGY_SURFACE_GEOMETRIC_HEIGHT
Surface Geometric Height (meters)
ASM PHIS
SKCLIMATOLOGY_SURFACE_PRESSURE_PA
Surface Pressure
ASM PS
SKCLIMATOLOGY_CLOUD_FRACTION
Cloud fraction
ASM CLOUD
SKCLIMATOLOGY_QI_MMR
Mass fraction of cloud ice water
ASM QI
SKCLIMATOLOGY_QL_MMR
Mass fraction of cloud liquid water
ASM QL
SKCLIMATOLOGY_QV
Specific Humidity
ASM QV
SKCLIMATOLOGY_RH
Relative Humidity
ASM RH
SKCLIMATOLOGY_AOA_DAYS
Age of Air in Days
ASM AOA
SKCLIMATOLOGY_DUST_0p7mu
Dust Aerosol, Reff 0.7 um
TRCDST1
SKCLIMATOLOGY_DUST_1p4mu
Dust Aerosol, Reff 1.4 um
TRCDST2
SKCLIMATOLOGY_DUST_2p4mu
Dust Aerosol, Reff 2.4 um
TRCDST3
SKCLIMATOLOGY_DUST_4p5mu
Dust Aerosol, Reff 4.5 um
TRCDST4
SKCLIMATOLOGY_BCPI
Geos Chem Black Carbon I
TRCBCPI
SKCLIMATOLOGY_BCPO
Geos Chem Black Carbon O
TRCBCPO
SKCLIMATOLOGY_SALA
Geos Chem Sea Salt aerosol Accum
TRCSALA
SKCLIMATOLOGY_SALC
Geos Chem Sea Salt aerosol Coarse
TRCSALC
SKCLIMATOLOGY_OCPI
Geos Chem Organic Carbon Aerosol I
TRCOCPI
SKCLIMATOLOGY_OCPO
Geos Chem Organic Carbon Aerosol O
TRCOCPO
SKCLIMATOLOGY_BRO_CM3
BrO
TRCBrO
SKCLIMATOLOGY_BRO_VMR
BrO
TRCBrO
SKCLIMATOLOGY_C3H6O_CM3
C3H6O. Acetone
TRCACET
SKCLIMATOLOGY_C3H6O_VMR
C3H6O. Acetone
TRCACET
SKCLIMATOLOGY_CH3I_CM3
CH3I. Methyl iodide
ASM CH3I
SKCLIMATOLOGY_CH3I_VMR
CH3I. Methyl iodide
ASM CH3I
SKCLIMATOLOGY_C5H8_CM3
C5H8. Isoprene
TRCISOP
SKCLIMATOLOGY_C5H8_VMR
C5H8. Isoprene
TRCISOP
SKCLIMATOLOGY_CH2O_CM3
CH2O. Formaldehyde
TRCCH2O
SKCLIMATOLOGY_CH2O_VMR
CH2O. Formaldehyde
TRCCH2O
SKCLIMATOLOGY_CH4_CM3
CH4. Methane
TRCCH4
SKCLIMATOLOGY_CH4_VMR
CH4. Methane
TRCCH4
SKCLIMATOLOGY_CO_CM3
CO
TRCCO
SKCLIMATOLOGY_CO_VMR
CO
TRCCO
SKCLIMATOLOGY_HNO2_CM3
HNO2. Nitrous Acid
TRCHNO2
SKCLIMATOLOGY_HNO2_VMR
HNO2. Nitrous Acid
TRCHNO2
SKCLIMATOLOGY_HNO3_CM3
HNO3. Nitric Acid
TRCHNO3
SKCLIMATOLOGY_HNO3_VMR
HNO3. Nitric Acid
TRCHNO3
SKCLIMATOLOGY_NH3_CM3
NH3. Ammonia
TRCNH3
SKCLIMATOLOGY_NH3_VMR
NH3. Ammonia
TRCNH3
SKCLIMATOLOGY_NITS
Inorganic Nitrates
TRCNIT
SKCLIMATOLOGY_NO2_CM3
NO2
TRCNO2
SKCLIMATOLOGY_NO2_VMR
NO2
TRCNO2
SKCLIMATOLOGY_NO_CM3
NO
TRCNO
SKCLIMATOLOGY_NO_VMR
NO
TRCNO
SKCLIMATOLOGY_O2_CM3
O2. Oxygen
ASM PL and T
SKCLIMATOLOGY_O2_O2_CM6
(O2)2for CIA
ASM PL and T
SKCLIMATOLOGY_O3_CM3
O3. Ozone
TRCO3
SKCLIMATOLOGY_O3_VMR
O3. Ozone
TRCO3
SKCLIMATOLOGY_PAN_CM3
PAN, Peroxy acetyl nitrate
TRCPAN
SKCLIMATOLOGY_PAN_VMR
PAN, Peroxy acetyl nitrate
TRCPAN
SKCLIMATOLOGY_SO2_CM3
SO2
TRCSO2
SKCLIMATOLOGY_SO2_VMR
SO2
TRCSO2
SKCLIMATOLOGY_SO4_CM3
SO4
TRCSO4
SKCLIMATOLOGY_SO4_VMR
SO4
TRCSO4
Modifying the storage location of GEOS CHEM data files¶
The storage location of GEOS CHEM files is stored in a python configuration file specific to each python environment. The value can be viewed or modified using a simple script installed with the sasktran_gcm module. Follow the example below:
>>> import sasktran_gcm.update_settings
>>> sasktran_gcm.update_settings.modify()
Package options for the Sasktran GCM Components
-------------------------------------------------
1) folder for the GEOS-CHEM database -> \\datastore\valhalla\root\data_products\G5NR_HERACLES
0) Save and Exit
-1) Quit and do not save
The default value of the storage location parameter is \datastore\valhalla\root\data_products\G5NR_HERACLES. It is appropriate for Windows machines located on the University Of Saskatchewan campus.
Data Files¶
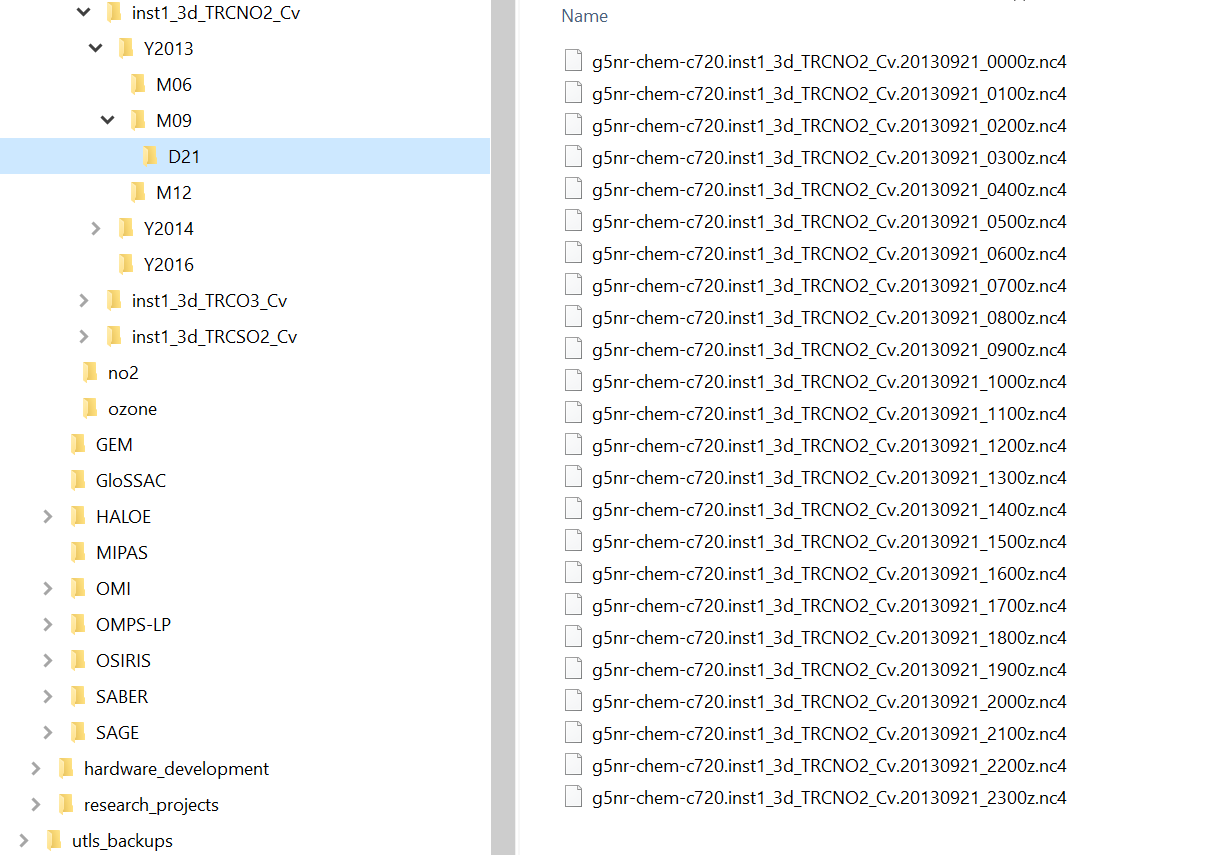
Each GEOS-CHEM variable is archived in sub-folders of the NCCS online GEOS-CHEM data portal and must be downloaded to the user’s system befor ethey can be used. The archive has a relatively complex directory hierarchy where each Geos-Chem variable has its own sub-directory tree of the format inst1_3d_xxxxxxx_Cv, where xxxxxx is the GEOS-CHEM variable name taken from the table above, eg TRCNO2. The directory hierarchy is organized by year, month and day which must be replicated on the user’s system. Geos-Chem variable data files are stored in the day folder at 1 hour resolution, see figure below. Users only need to download the files they wish to use in their simulation. Utilities such as curl and wget may prove useful for the download task. Keep in mind that the Sasktran Geos-Chem Climatology always uses the two files that straddle the UTC time of interest and you both files need to be on the local system.
Other Geos-Chem variables, most notably pressure and temperature, are taken from the GEOS-CHEM assimilation files which are stored in a slightly differently format under the inst1_3d_asm_Cv folder. These variables are marked in the above table with ASM in the GEOS-CHEM variable name column. The directory hierarchy is again organized by year, month and day and the assimilation data files are stored in the day folder at 3 hour resolution compared to 1 hour for the regular variables, see figure below. All assimilation variables are stored in the same assimilation file.
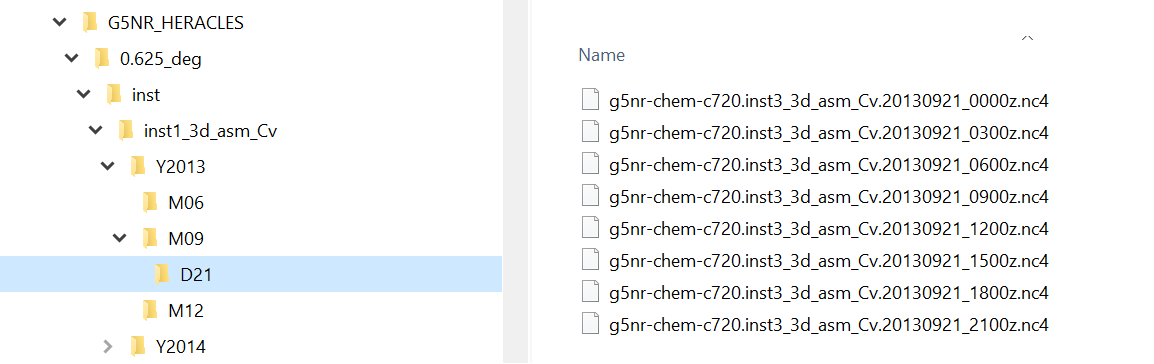
User’s will always need to download the two assimilation files that straddle the UTC time of interest of their simulation as these files contain several mandatory parameters, e.g. geometric height of the layers, surface geopotential, pressure and temperature.
Downloading GEOS-CHEM from NCCS DataPortal¶
You can use wget to download data from the NCCS dataportal and retain the directory hierarchy. For example, the following will download the assimilation data for 2014-03-21,:
wget --accept *.nc4 -r -np https://portal.nccs.nasa.gov/datashare/G5NR-Chem/Heracles/12.5km/DATA/0.625_deg/inst/inst3_3d_asm_Cv/Y2014/M03/D21/
The key features of the wget command are
–accept *.nc4 to only fetch the netcdf4 data files
-r to be recursive and retain directory structure
-np to exclude parent directories, otherwise you get the whole site.
There is also the option to use –directory-prefix to specify the output directory location.
Interpolation¶
The GEOS-CHEM sasktran package performs interpolation with the following steps,
The four grid points that surround a given location are calculated and accounts for any longitude wraparound.
Height profiles of each required variable are generated for each of the four grid points for each time that straddles the point of interest. The code internally accounts for the geos-chem variables on a 1 hour grid and the assimilation variables on a 3 hour grid.
Values at the height of interested are generated for each of two sets of 4 vertical profiles using the interpolation techniques assigned to the variable.
Horizontal linear interpolation is applied to each set of 4 vertical profiles, linearly interpolating in longitude and latitude.
Linear temporal interpolation is applied between the two sets of vertical profiles.
Vertical Extrapolation¶
This still needs to be worked out. The current version sets most values to zero below the topographic surface and above the highest value.
Vertical Interpolation¶
This still needs to be worked out. Most variables are linearly interpolated. Air number density and pressure are log interpolated.